Synthetic Origins

Introduction
In 1997, as Michael Elowitz was studying for a Ph.D. in physics at Princeton University, he had a remarkable idea: What if it were possible to build a circuit out of DNA, rather than electronics, and use it to “program” a living cell?
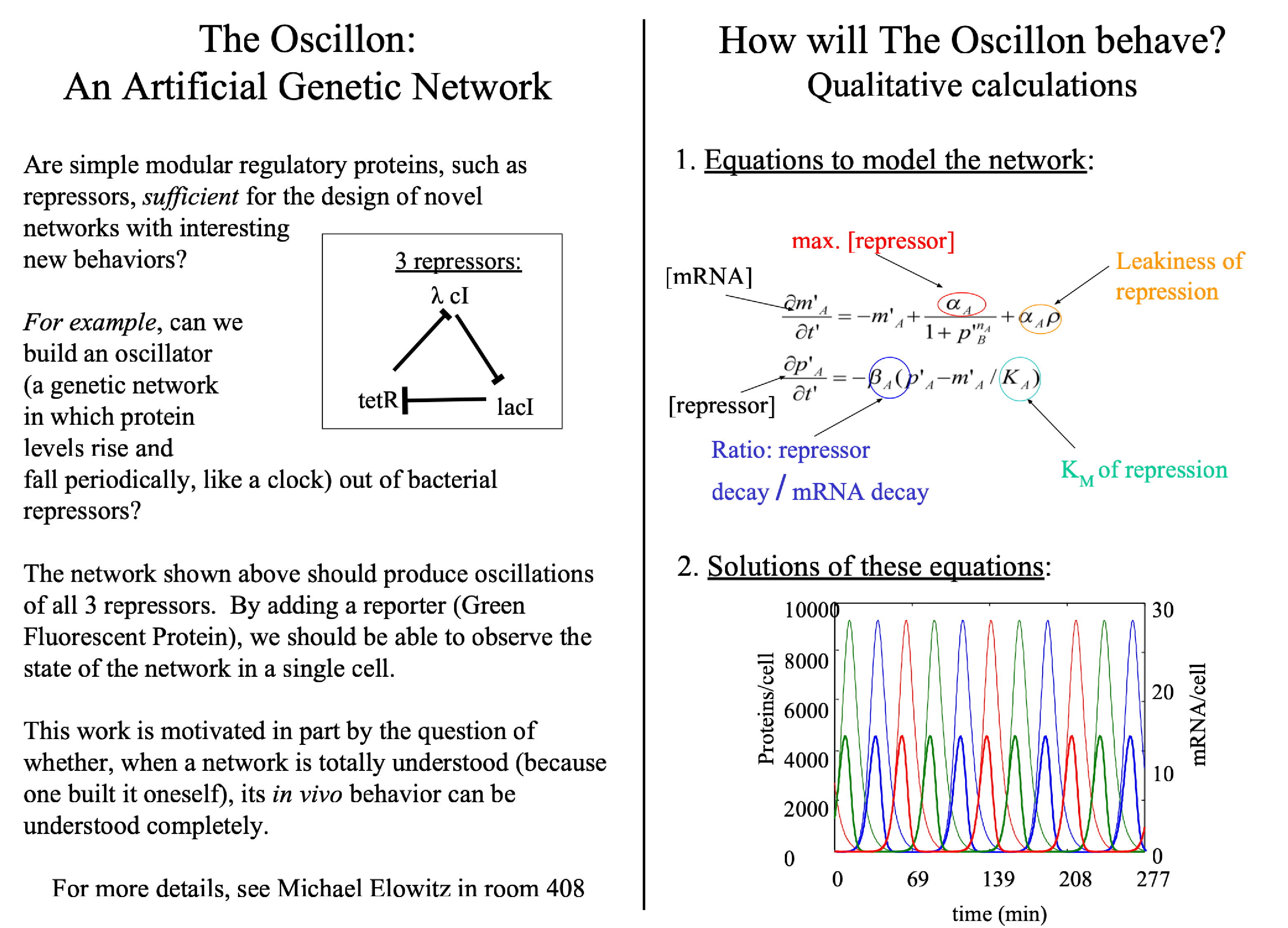
For the next three years, working with advisor Stanislas Leibler, Elowitz labored over this synthetic gene circuit and achieved precisely that. His final design, now known as the repressilator, was made from just three genes, each of which encoded a repressor protein that blocks transcription of the next gene in the loop. For example, LacI represses the promoter controlling the expression of TetR, TetR represses the promoter controlling the expression of cI, and cI represses the promoter controlling the expression of LacI. A gene encoding green fluorescent protein was placed alongside one of the three promoters, and as the engineered cells cycled from one gene to the next, they blinked between green—dark—green—dark. Elowitz had created cells that acted as living oscillators.
The repressilator was published in Nature in 2000, back-to-back with another paper that reported a second kind of synthetic gene circuit, called the toggle switch. These papers became the standard-bearers for a new scientific field: synthetic biology.
Together, these early synthetic gene circuits suggested that engineers really could recreate some of the complex networks within living cells and then manipulate them to carry out entirely new functions. In other words, they could “program biology.”
Elowitz, now a professor of biology, bioengineering, and applied physics at the California Institute of Technology, recently sat down for an interview with Asimov Press. During our conversation, he reflected on his early doubts while working on the repressilator, mused on the unique philosophies that physicists bring to biology, and highlighted his group’s recent projects, published and unpublished, in building technologies to decode the mechanisms of living cells.
This interview has been edited for length and clarity.

Interview
Asimov Press: I’m a bit embarrassed to start this interview by asking you about the repressilator. Everybody must ask you about it. But I noticed one thing when I looked at your paper side-by-side with the toggle switch paper that I wanted to ask you about. And that’s that your discussion section focused on the importance of synthetic gene circuits to interrogate natural biological systems, whereas the other paper focused more on the commercial potential. Can you expand on that?
Michael Elowitz: Well, my motivation for practicing synthetic biology was often more on the foundational side, in using it as a way to explore how cells work. But I also always wanted to get at this question of whether we can really program a cell the way we program a computer. That was just a really attractive idea.
But in those early years, everything in the laboratory was so slow and difficult. Even building the repressilator, which only has three genes and seems trivial by today’s standards, took me years to build. I had to characterize each transcription unit, and I tried to set it up in a way where I could measure each transfer function from one repressor to the next independently, and then try to concatenate them into two gene steps, and so on.
AP: The synthetic biology field took off shortly after that issue of Nature in 2000. What did you make of its rapid growth?
Elowitz: Initially, there was this period dominated by a small but growing handful of labs taking baby steps and implementing new biological functions, one by one: digital logic, computation, and quorum sensing. These small steps allowed more sophisticated capabilities later on.
I always believed in the vision of programming cells, though, and felt like there were more fundamental things we’d have to know to achieve that vision. To make our circuits function and allow them to use core cellular capabilities, we would have to learn the API of the various cellular systems—transcription, signaling, and the cell cycle—so to speak. And we would need to figure out basic aspects of system behavior that had not been measured quantitatively or at the single cell level.
For example, shortly after the repressilator paper, I worked on another paper to measure a simple response function—the quantitative effect of a repressor on its target gene. This is the elementary transfer function whose shape and fluctuations are critical for predicting how larger transcriptional circuits will behave. That was a collaboration with Nitzan Rosenfeld, Jon Young, Uri Alon, and Peter Swain.
At the same time, while these real—and to me very exciting—experiments were happening and interesting papers were being published by a small but growing number of labs, synthetic biology became really hyped up. And that hype seemed to exceed where the field actually was. Because of my own anxious personality, I worried at the time that that excessive hype would come back to bite us all in the tuchus if we, as a field, made promises that we couldn’t possibly keep within a reasonable timescale. In retrospect, this worrying was unnecessary.
{{signup}}
AP: Can you elaborate on what you mean by the API of a cell?
Elowitz: I mean the biochemical interfaces through which natural cellular functions can be controlled by synthetic circuits. Transcription and translation are the most obvious examples since they can be used to express any gene of interest. But the capabilities of synthetic circuits depend on their ability to control as many endogenous functions as possible: the cell cycle, growth, morphological states, communication with other cells, death programs, export of cellular components, and so forth. It is often unclear, though, how these cellular systems can actually be controlled in a specific manner, and in a way that doesn’t also mess with other processes, by a synthetic circuit. So discovering these control points—defining the cell’s natural APIs—is a really important aspect of synthetic biology.
AP: Where did the idea for the repressilator first come from?
Elowitz: It must have been sometime in 1996 or 1997. I was a graduate student in Stanislas Leibler's lab at Princeton. A lot of people there were trying to make models of different biological circuits, such as chemotaxis. I was reading a lot of molecular biology papers about different systems, especially circadian clocks, and also a book called Biological Feedback, by Thomas and D’Ari, that sketched out really simple models for different kinds of feedback circuits.
And as I’m reading all of these papers, I noticed that many of them concluded with a cartoon model of the biological circuit deduced from the genetic or biochemical measurements in the paper. What I most remember is just feeling like, are these circuit sketches really sufficient to explain the behavior? Or are they just a summary of observed interactions, possibly omitting many other critical components? It was driving me crazy.
Like, how could someone ever know what set of interactions and components are sufficient to explain a cellular behavior? When you approach a circuit from the top down, you can’t know for sure you're not leaving out components. But if you build the circuit up from scratch, out of well-defined components and interactions, then you could figure out whether a circuit was actually sufficient for a behavior.
So that was the idea. I wanted to just build a gene circuit and see if I could get it to carry out a specific behavior. The repressilator came from this high-level goal. But the reasons I focused on an oscillator over other circuits had to do with all the oscillators I studied in physics, the circadian clock I had been reading about, and above all the idea that I could make cool movies of blinking bacteria. I just thought it would be so cool to build a gene circuit that could blink.
AP: Was that always the name—the repressilator?
Elowitz: Well, at least up until 1999, I called it the oscillon, which was a portmanteau between the words oscillator and operon. But one day I saw an issue of Nature with the word oscillon on the cover. I think it said, ‘Oscillons observed!’ or something like that. The oscillon, in that case, referred to a coherent oscillatory mode of granular media. And then I was like, ‘Oh dang, they took it.’ But I’m glad they did, because the repressilator turned out to be a better name. Besides being fun to say, it also connects to a tradition of nonlinear dynamic models of chemical oscillators named for the cities where they were conceived: the Brusselator, the Palo Altonator, and so on.
AP: Back when you were trying to build the repressilator, what were your thoughts on the project—and what were people around you saying about it?
Elowitz: Well, I definitely had no idea whether it was going to work. When I asked people what they thought of the project, which I did incessantly, I got very different answers. A few well-known biologists would say, ‘No, it’ll never work that way. It just won’t work.’
And I’d ask them, ‘Why won’t it work?’ And they’d say, ‘Biology just doesn’t really work that way. You can’t predict what’s going to happen.’ Other people thought it sounded fun. So it was a mix of positive and negative feedback. It’s funny to think about that in hindsight. At the time, I was really excited about the project. I told lots of people about it, but then I’d swear them to secrecy. It was all very silly.
At the same time, my advisor, Stanislas Leibler, who is a truly visionary scientist and a personal inspiration, was supportive of the project throughout, which is something I have always really appreciated.
I was actually building the repressilator during Stan’s sabbatical year at the EMBL in Heidelberg, Germany in 1998. During that year, I was looking for the best gene regulatory components I could find. Hermann Bujard had done careful work analyzing the features of bacterial promoters that could allow them to be both strong and tightly regulated, two of the key properties necessary for the repressilator to work. And he was also in Heidelberg. So Stan and I went down the hill to chat with him, and we showed him the repressilator design. Bujard looked at our design, mentioned that he had always wanted to build something else, and proceeded to draw a toggle circuit!
AP: Wow, what a coincidence. When you finally published the repressilator in 2000, were you aware that it would be printed side-by-side with the toggle switch made by Timothy Gardner, Charles Cantor, and Jim Collins at Boston University? Presumably, you thought you were the only one building a synthetic gene circuit.
Elowitz: Yeah, I did not know anyone was working on the toggle switch until I literally opened a copy of the journal.
AP: When you finally got the repressilator working, did the meaning of the moment sink in? Did you know that you were inventing this whole new field of biology?
Elowitz: Well, nobody at Princeton was telling me that if I did this thing, it was going to be amazing. I was not getting that feedback at all. And I never really thought of it in those terms. I just thought it would be really cool. I genuinely wanted to know if it could work, and I kept envisioning the movie that would show it in action.
After the paper was published, I gave a bunch of talks about it, and I think there were positive reactions, but it wasn’t until a few years later that there was really an inflection in the field of synthetic biology, around the mid-2000s.
AP: After you published the repressilator, I heard that it was difficult for others to replicate the results; it was a bit finicky to get working. Is that true?
Elowitz: I don’t think that’s quite right. In the paper, though, we emphasized variability in the dynamics. And there was an increase in basal fluorescence over time. And that's because the conditions I was using, where I was putting the cells on an agarose pad, were not steady-state. So over time, waste products pile up in the media, and the cells grow more slowly.
Years later, Laurent Potvin-Trottier, Johan Paulsson, and colleagues took the original repressilator and put engineered cells in a microfluidic channel, in a Mother Machine, which allowed them to follow the dynamics over hundreds of cell generations in constant conditions. And the repressilator oscillated forever, basically. But there was a lot of variability, for sure, that Laurent and Johann later explained in their paper.
AP: What was the explanation?
Elowitz: Well, it’s kind of counterintuitive. The original repressilator included three repressors, where each one represses the next. And one of the repressors was tetR, which binds very tightly to its operator. Johan showed that because the binding is so tight, the end of the tetR phase of the oscillation requires tetR to go away enough to de-repress its target.
But because its binding is so tight, that requires waiting until the last molecules of tetR dilute out of the cell, which makes it very sensitive to stochastic fluctuations. And that's actually one of the biggest sources of variability in the repressilator’s dynamics.
So Johan fixed that and made a version that's astonishingly precise and reliably oscillates for much longer, retaining its phase for 180 cell generations or so.
AP: What did you think about the term ‘synthetic biology’ when people started using it? And do you feel like the field has reached an inflection point now, where a lot of the hype you initially worried about is now becoming real?
Elowitz: I always liked the term. It really fits perfectly, and it is analogous to chemistry, where you've got synthetic chemistry and analytical chemistry. And in terms of the field, yeah, I do. The really crazy stuff is still out of reach, but the basic premise that synthetic biology will enable you to improve or create useful therapeutics, for example, that actually take advantage of the ability to program the cell, is slowly becoming real. And that’s exciting. Everything took a lot longer than anybody expected. But the fundamental premise that the cell is—to a large extent—a programmable device, appears to be valid.
AP: Is the cell a computer? Or is there a better analogy?
Elowitz: The computer analogy is a double-edged sword.
The analogy is useful in the sense that a cell is a programmable device that can do many different things. And the closest thing we have like that, in our daily life, is the computer. You can program your computer to do all kinds of things. And I think that's also true with the cell; you can program the cell to grow, divide, change morphology, interact with other cells, and do all kinds of things that are difficult for us to imagine. Cells are open-ended, programmable systems.
We can even program cells to carry out functions that cells did not naturally evolve to do. So from that point of view, the computer analogy seems quite useful and accurate, actually.
But there's also a lot of ways in which a cell is not a computer. And I think those are equally important. Cells are noisy, and they use that noise to control behaviors at the population level. They also make copies of themselves and grow exponentially—computers do not do that. Instead of transistors connected by wires, cells use molecules connected by specific molecular interactions.
Another one is negative numbers, right? You can't have a negative concentration of a molecule in biology, which means that biology has to solve problems in unique ways. There’s also combinatoriality, in which biological systems encode signals in combinations of molecules that compete to form different complexes. These systems compute in ways that resemble digital computation in some ways and differ in others.
So the thing that makes me nervous about the metaphor is when we start to impose our electrical engineering expectations on the mysterious world inside of a cell. There are some principles of electrical engineering that apply to living cells, but the most interesting things about biology are all the ways in which they’re different.
AP: I recently spoke with Ed Boyden, and he said something that really resonated: ‘Understand before you augment.’ I’m curious if you think there are tools we should build to understand cells at a greater depth, such that they become less mysterious. What things do we need to figure out before we can actually program cells reliably?
Elowitz: That’s a great question. I think that there are so many different things. I mean, we’re getting better at understanding how cells control genes, but we don’t even have the ability to rationally program a gene to be expressed in all types of cell.
AP: We can’t even grow most types of cells in the laboratory.
Elowitz: Exactly. We also can’t track the activities of all the kinases in a cell, or anything like that. So if you think about all the different levels of information and states in a cell, its spatial and temporal organization, and compare that to what we can currently read out and manipulate, it's actually quite primitive.
At the systems level, delivery is a big challenge. We can design synthetic gene circuits that work perfectly in a petri dish, but it’s still difficult to get those genes into cells. So I think that's a hugely important area. Another one is immunogenicity. Like, if we start thinking about therapeutic circuits, how can we design the components in such a way that they are not going to cause an immune response?
AP: It seems like your lab is trying to build tools and technologies that fill in some of these gaps. But do all the things that we don’t know about biology actually inform which tools your lab decides to build? Or do you just follow your creative interests?
Elowitz: It’s a mix. A lot of times we make tools because we imagine something and just really want it to exist. The RNA Exporter, which was developed by Felix Horns and others in my lab, is one example. It came from the idea that everyone studies RNA and it’s useful to look at the transcriptome and how it changes over time, but these tools all require that you kill the cell. But we wanted to know what's going on dynamically, and how we might track it. So we engineered a system that allows the cell to package up RNAs and then secrete them in extracellular vesicles. By sequencing the RNAs that come out of the cells, you can track cell dynamics non-destructively, without killing the cells.
The RNA Exporter also led to another application, which is related to the delivery issue. We have always wanted to find better ways to get synthetic gene circuits into cells so we could use them as therapeutics. We wondered, what is the optimal way to do that? And we figured that the best delivery vehicle to get something into a cell is to use another cell.
We began to think, okay, what if we could just program a cell to home to a specific part of the body, recognize a specific cell located there, and then get this ‘delivery cell’ to switch on and start exporting RNA at that location? The exported RNA could transfect just the target cell.
So the RNA Exporter provides a way not only to track RNA molecules over time, but also a potential way to deliver RNA as well.
AP: It’s interesting that your work begins with these big questions of imagination, and you use that to slowly work your way toward useful, but out-of-the-box, technologies.
Elowitz: Yeah. I mean, of course, we could just work with existing tools and focus on building biological circuits that way. But I feel like whenever you expand the toolbox in synthetic biology, it takes you to some interesting places. You can't do everything, obviously, so you do need to pick which tools to focus on. A lot of the time, my interests go to things that are mechanistically or aesthetically beautiful: elegant solutions that are going to help us get to a new kind of biology.
AP: Last question: Why have physicists so often been amongst the most important biologists throughout history?
Elowitz: Naivete? Arrogance? A belief in the understandability of nature? I don’t know. There are so many great examples of physicists coming into biology. But I think we should be careful with that. So many kinds of people have contributed to biology in different ways.
AP: But is it a coincidence that physicists were so important at the start of molecular biology, or that physicists, yourself included, kicked off the field of synthetic biology?
Elowitz: I think there's something about the physics style of reasoning, thinking, and problem solving. Physicists value reducing a problem to its essence and imagining the simplest example of a question and the simplest way to go after it. They also value quantitative approaches. If you think about the initial synthetic gene circuits, that's exactly what they were. They're literally the simplest, non-trivial circuits anyone could imagine.
But in biology, we continuously discover that biological systems are far more complicated than we realize. Biologists find the TGF beta pathway and then discover all these modulators and all these modulators that modulate the other modulators. So biology is a kind of never-ending process of uncovering and unpacking all this complexity.
The synthetic approach—building to understand—is really just saying that scientists should start with the simplest possible circuits and see what they can do and build up from there. I think that’s a style that's appealing to physicists, and certainly was appealing to me. But the two approaches—synthetic biology and ‘real’ biology—are complementary. We don’t want a world with only one.
With a question like, ‘How does an oscillator within a cell work?’ you need to come at it from both directions. You need to have people who map out the components of a natural oscillator and figure out how they interact. But then it helps to also have someone say, ‘Okay, but why do we need that complexity? What is the minimal circuit that can generate this behavior? How susceptible will that minimal circuit be to noise or temperature? How can it operate more robustly? Are there other compensating mechanisms outside the oscillator in the cell that are making it robust?’
Often the only way to find out how something really works is to control the interactions yourself. It’s to go build the thing and then watch it work. I think that impulse has been around for a long time in biology, but we just didn’t have the basic genetic engineering tools that we needed to enable it until recently.
{{divider}}
Cite: Michael Elowitz. "Synthetic Origins." Asimov Press (2024). DOI: https://doi.org/10.62211/21yr-22te
This article was published on April 2, 2024.
Always free. No ads. Richly storied.
Always free. No ads. Richly storied.
Always free. No ads. Richly storied.