Inside the Laboratory for Extraordinary Microbes
E. coli, the most studied microbe of all time, was first discovered in baby poop by German pediatrician Theodor Escherich in 1885. He had been isolating microbes from newborn baby feces to study the effects of breastfeeding on gut health. He originally called the cells “bacterium coli commune,” though the name was later changed to Escherichia coli in his honor.
Before World War II, few scientists used E. coli in their experiments. But in the 1950s, at the dawn of molecular biology, scientists gained a new appreciation for it after searching for organisms that were easy to work with and quick to grow. Escherich’s microbe, which divides in just 20 minutes and can thrive in a plethora of liquids, fit the bill.
In 1952, when the famous Hershey-Chase experiment indicated that DNA—and not protein—was the source of genetic material, just 285 papers mentioned E. coli. But the bacterium’s prevalence swelled from there. Last year, more than 14,000 articles reported the use of E. coli, and the actual number is likely far higher.1 This singular organism has become the de facto microbe for molecular biology.
But what are the odds that a microbe discovered in baby poop in 1885 is actually the best organism with which to advance scientific progress? Probably slim. Humanity has discovered an estimated 0.001 percent of all microbes, and many of biology’s most useful tools have come from the “weird” ones.
Consider that scientists found one of the first restriction enzymes, used to “cut” and “stitch” DNA molecules together, in a pathogenic bacterium hiding out in the respiratory tracts of children, called Haemophilus influenzae. Alexander Fleming, upon returning from holiday, discovered penicillin antibiotics after a little-known mold destroyed his Streptococcus colonies. And an enzyme isolated from a microbe growing in the boiling waters of a Yellowstone National Park geyser enabled modern polymerase chain reaction, or PCR.
Scientists don’t often work with “non-model” microbes because they are, simply, more difficult to handle than E. coli, on which many molecular biology tools are tested, refined, and perfected. A myopic focus on E. coli has trapped molecular biologists in a ditch from which it is hard to escape.
But a small group of scientists at Cultivarium—”a non-profit that aims to democratize access to extraordinary microbes”—are working to change that. Cultivarium builds tools to grow, transform, and engineer “extraordinary” microbes: the salt lovers, heat tolerators, and geyser growers that have long been inaccessible to scientists. Studying such organisms might lead to better gene-editing tools or medicines. And regardless of whether Cultivarium’s engineering efforts succeed or fail, they give all their knowledge away for free, in the expectation that the next biological breakthrough will come from these overlooked microbes.

Henry Lee, Cultivarium’s CEO, has an affable face and a charming tendency to speak his mind, traits that came in handy when he had the idea to start the nonprofit about a decade ago. At the time, he was a postdoctoral researcher at Harvard University, studying a peculiar microbe called Vibrio natriegens, which divides in less than ten minutes, or twice as fast as E. coli.2
After five years of work, Lee published a paper that reported the first molecular tools used to grow, transform, and engineer V. natriegens. He also produced the first complete genome sequence for the organism and, more importantly, found his life partner in the process.
Nili Ostrov is a fast-talking Israeli scientist with piercing green eyes and an intensity that compliments Henry's calm demeanor. After a few years working together at Harvard, the duo wondered whether the work that had taken Lee six-and-a-half years to complete for Vibrio could be replicated in weeks, or even days, for other organisms.
The pair now run Cultivarium together, says Ostrov, because “it just felt like we could solve a fundamental, but neglected problem.”
{{signup}}
Microbe House
Cultivarium nestles on the first floor of a two-story, steel building in Watertown, Massachusetts. Lee showed me around the laboratories on a clear day in April. After we donned disposable lab coats, he ushered me toward four rows of research benches equipped with standard tools of the trade—micropipettes and centrifuges. Bright light streamed through tall windows. I peeked into several rooms and spotted a great variety of large (and often proprietary) machines.
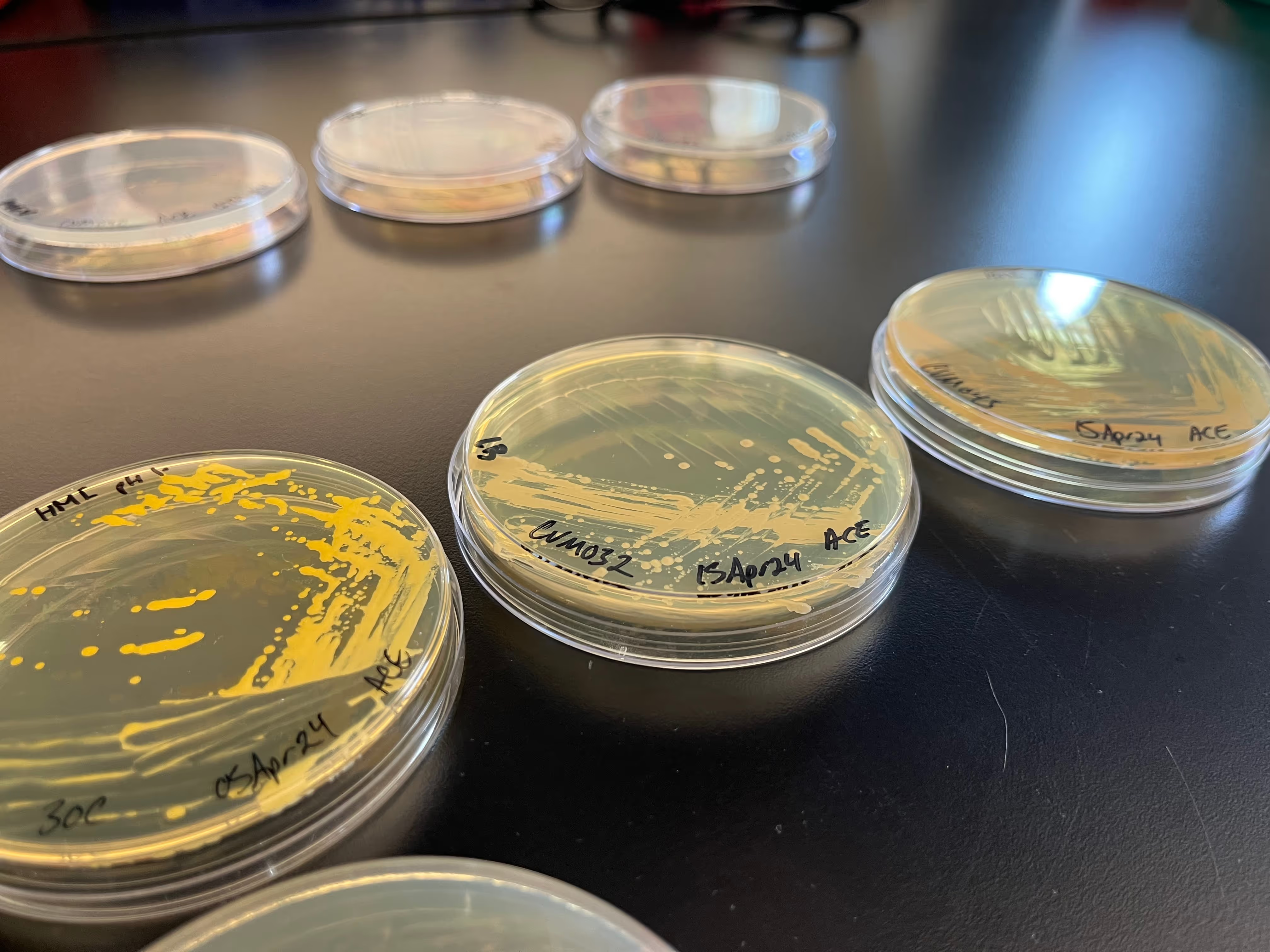
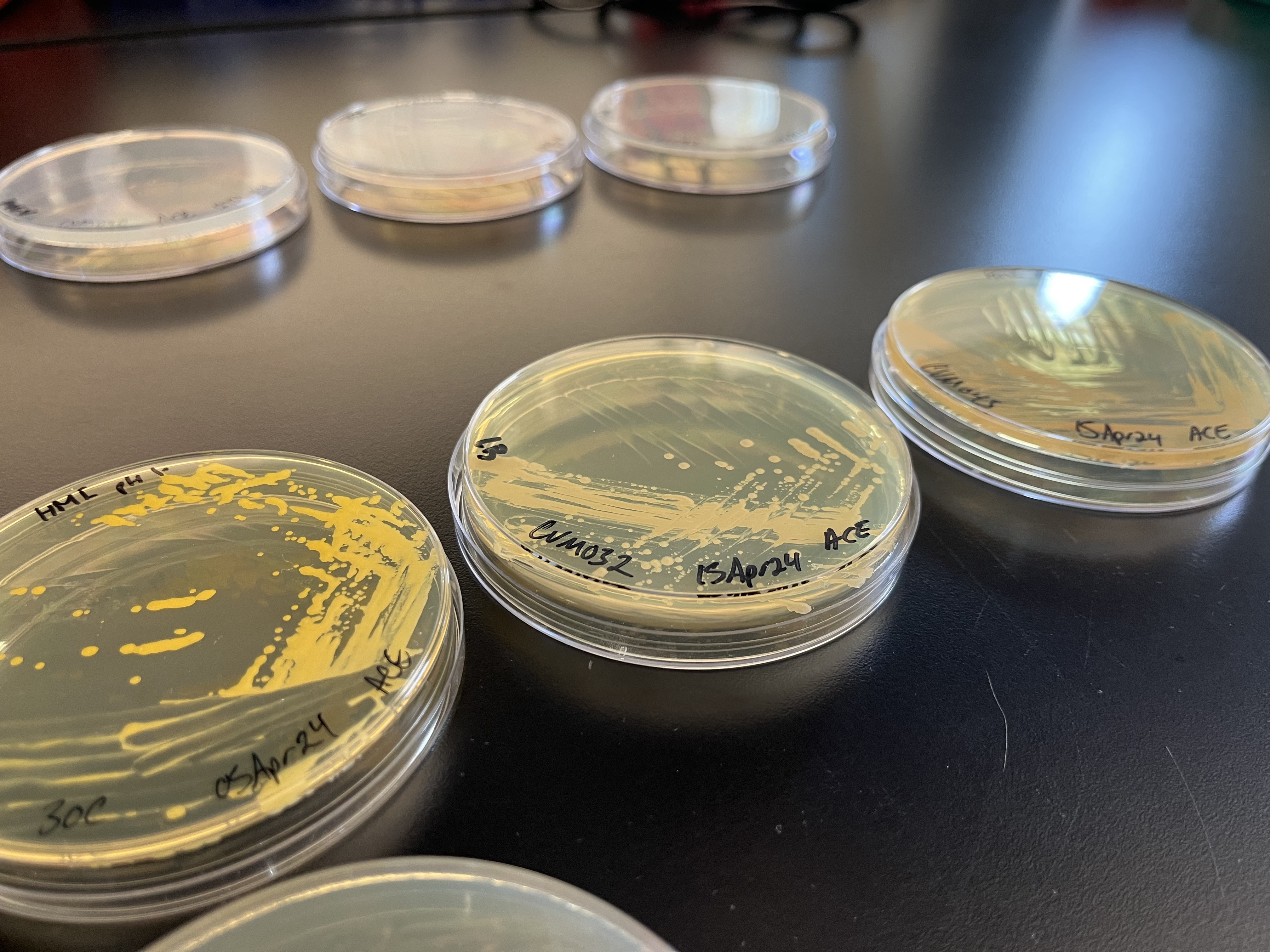
While the laboratory itself wasn’t out of the ordinary, some of its contents were. At one of the benches, Lee pointed to several agar plates that had been streaked with different non-model microbes. An alkiphile—an organism that thrives in alkaline environments around pH 10—bloomed rosily. Nearby, a halophile, or salt-loving microbe, emitted a bright yellow hue.
As we walked, Lee told me that Cultivarium’s efforts to make “extraordinary” organisms accessible almost always follow the same basic steps. First, the team orders a microbe from ATCC, a non-profit group that has been storing and mailing microbes to researchers since 1925. The ATCC catalog includes more than 14,000 bacterial strains, the vast majority of which gather dust and are rarely ordered by researchers.
After receiving a microbe in the mail, Cultivarium sequences it. Mutations can creep into strains over time, and even a seemingly minor alteration—a single base swapped here or there—can change how cells grow and respond to their environment.
Lee told me that he once sequenced Vibrio natriegens stored in the ATCC database. Ten years later, a professor at Harvard ordered the same microbe from ATCC and sequenced its genome again. But the professor noticed a small change: the Vibrio cells now carried a single mutation in a ribosomal gene that made the cells sickly and slow-growing. This mutation had not been present when Lee studied the same microbes just a decade prior: evidence that nothing in biology remains constant. By sequencing the genome, Cultivarium constructs a record from which to diagnose future problems.

Next, Cultivarium grows the microbes, which can prove a difficult task. Some cells thrive at high temperatures, low pH, or high acidity. Some cells need particular amino acids or salts simply to live, let alone to thrive. Other microbes have exotic nutritional needs, such as requiring mucin, a protein found in human mucus, for their survival. Many of these parameters are difficult to predict ahead of time, especially when an organism has never been cultured before.
The trial-and-error nature of microbial cultivation has not changed much in the last century-and-a-half. In 1860, Louis Pasteur brewed the first liquid artificial culture medium. His initial concoction consisted of a “yeast soup,” made by crushing cells with a mortar and pestle and mixing them with ash, candy sugar, and ammonium salts. He aimed to assemble a broth with all the elements needed to support growth; ammonium salts for nitrogen, sugar for carbon, and ash for vitamins. Microbes added to this mixture sometimes grew, but more often didn’t.
In 1881, Robert Koch showed that cells grew better when incubated “in a broth composed of fresh beef serum or meat extract.” He used this recipe, and modifications thereof, to discover the microbes that cause tuberculosis, cholera, and anthrax. In 1882, Koch also demonstrated that adding agar to the extracted liquid formed a gel on which cells could be streaked and isolated.3
These early microbiologists spent years fine-tuning their concoctions. Today, Cultivarium solves these problems with robots.
In a small room called the “Growth Chamber,” Cultivarium uses a liquid-handling robot to dip microbes into hundreds of little wells etched into a plastic plate. Each well contains a unique growth media—some made from blood and others containing mucin, for example. A camera mounted to the robot takes photographs of each well at recurring intervals, tracking which ones become cloudy (indicating that the microbes are growing) and which wells remain translucent.4
Growth is only the first step in working with a microbe, however. Lab-grown cells can be studied beneath a microscope or battered with antibiotics to see how they fare. But most discoveries today are made by transforming cells—literally coaxing microbes to take up foreign DNA—to add, remove, or edit genes. Transformation is the linchpin of modern biology; the tool by which scientists link an organism’s genome to its phenotype, or visible behaviors and abilities.
In 1970, two biochemists at the University of Hawai’i figured out how to sneak DNA molecules into E. coli using calcium chloride. The calcium ions, which carry a positive charge, open tiny pores in the cell’s membrane while coating the negatively-charged DNA strands. In light of this ability, they act as a molecular lubricant to shuffle DNA past the negatively-charged cell membrane.
Another method to transform cells, called electroporation, was first described in 1982. That’s the year Eberhard Neumann at the Max Planck Institute for Biochemistry employed short “electric impulses” to punch holes into cells, allowing DNA to slip inside. Neumann demonstrated his method on mouse cells, but later showed that his technique transferred to lots of other lifeforms, too. Cultivarium uses electroporation to transform their microbes.
In the “Transformation Room,” Cultivarium uses a big, custom-built machine to shock cells. The machine cycles through voltage levels, waveforms, durations, and other parameters—out of millions of possible combinations—to find the “Goldilocks” zone to transform a particular microbe. There’s a fine line to walk here; too much voltage fries the cells, whereas too little means the DNA won’t have enough time to glide through the opened membranes.
Once Cultivarium finds a microbe’s preferred electroporation settings, they can unleash the full arsenal of modern genetics and genomics on studying and engineering these extraordinary bacteria. Cultivarium creates many different DNA sequences and floods all of them into microbes, simultaneously. Then, they observe which DNA strands make functional proteins. Over time, the scientists spot trends and come up with rules—this sequence made a protein, but this other sequence killed the cell entirely—to slowly, painstakingly, gain a deeper understanding of an organism. The team spends about 90 percent of their time on this step alone.
Over the last several years, Cultivarium has studied 106 organisms out of the 14,000+ stored in the ATCC catalog. This may seem like a small number, but each microbe requires an artisanal approach. Cultivarium works with the microbes that nobody else can, on the frontiers of scientific discovery, and often without anywhere or anyone to turn to for help.
“We’re fighting against organism loneliness,” Ostrov says, referring to their desire to expand the number of microbes available to scientists. Lee nods and says: “We’re all fighting against loneliness.”

Democratize Biology
Significant advances in science today do not often come from solitary geniuses, as they often did before World War II and the modern era of multi-billion dollar government programs. Much more commonly, progress stems from the collective efforts of large teams with aligned missions.
The Hubble Telescope and CERN both cost billions of dollars to build and required resources that far exceeded those available to an academic laboratory. Neither of these indispensable technologies turn the kind of profit that would attract venture capitalist funding, either. This is why scientific moonshots that could improve humanity, but which cost too much money to get up-and-running or are unlikely to return investments, often die at the idea stage. Focused research organizations (FROs)—Cultivarium included—are designed to fill that gap.
FROs tackle technical objectives that require resources greater than most academic laboratories can muster. They are not (at least initially) for-profit. Instead, FROs operate over a five-year period, make as much progress as they can, and then give away their findings or inventions to spur wider progress in a scientific field. This scientific structure is growing in popularity as people recognize the chasm between academic laboratories on the one hand and venture-backed startups on the other. Convergent Research, the non-profit organization that helped spin up Cultivarium and six other FROs, matches scientific proposals with potential funders who want to make them happen.
Cultivarium is at the midway point of their five years. With their remaining time, the team’s priority is to get all their findings and progress out into the world, mainly by posting them on a web portal. Their database already boasts 180,000 organisms, many of which have genome sequences. For the 106 microbes that Cultivarium has directly worked on, the database also lists data on their preferred growth conditions—temperatures, media, doubling times, plasmid origins for genetic engineering, and availability from strain banks.
As Cultivarium’s data swells, it also acts as a flywheel to drive future progress. The team now has enough data, for example, to train computational models that can predict optimal growth conditions for microbes, even when the microbe has never been cultured. The model ingests genome sequences and spits out predictions for a microbe’s desired oxygen, temperature, salinity (or saltiness) and pH levels.
The model was trained on nearly 16,000 bacteria and archaea for which there exist only partial genome sequences or growth data. Cultivarium left out entire clades—families of organisms evolved from a common ancestor—from the training dataset. They then used the model to predict growth requirements for those “untrained” microbes and benchmarked everything by growing those same microbes in the laboratory and measuring the results.

The model predicted oxygen tolerance with 92 percent accuracy and optimum temperatures with 73 percent accuracy. pH levels were more difficult to predict. But, in general, the model was able to make accurate predictions even when just 10 percent of a microbe’s genome sequence was available. In other words, the model can make a reasonable assumption about growth conditions for entirely new taxonomic groups of microbes even when just a tiny part of its genome has been sequenced.
This is great news for biology. Since the time of Koch and Pasteur, biologists have been myopic with their models (and it’s not entirely their fault!). As Cultivarium democratizes access to a broader palette of lifeforms, they hope that other researchers will continue their work, discover useful tools, and fuel the machines of progress.
“There are these moments in history where someone studies something weird and that catalyzes a step change,” says Lee, nodding to important discoveries like CRISPR gene-editing or the enzymes used in PCR. “I don’t know if I’ll ever be in that position to catalyze it,” Lee says, “but that’s a niche that I want to be in.”
{{divider}}
Niko McCarty is a founder of Asimov Press.
Thanks to Xander Balwit and Ethan Freedman for editing this essay.
Footnotes
1. These numbers come from PubMed. which includes about 37 million papers from the biomedical literature. Many journals—especially in China—are not included in the database.
2. Vibrio was first discovered in the salt marshes of Sapelo Island, off the coast of Georgia, in 1958. Nobody knows for sure why it divides so quickly, but a leading theory suggests that it has a large number of ribosomal RNA operons. This avoids one of the key bottlenecks limiting cell division: the time required to make ribosomes.
3. Anaerobic microbes, which can die if exposed to oxygen, were eventually cultured in 1915 using vacuum-sealed jars.
4. The team is so meticulous in their preparations of growth media that they even purify and quantify all the contaminants in the water used. When Cultivarium finds growth conditions that work for a particular microbe, they report the exact conditions—including water contaminants—in relevant documents. The brewing industry does something similar. When the Samuel Adams Brewing Company was first founded in 1984, they made beer using Boston tap water. And they do the same today—except their main manufacturing facility is in Pennsylvania. To maintain consistent taste across batches, and because it’d be inordinately expensive to ship water across state lines, Samuel Adams meticulously tests and quantifies the minerals and other compounds in Boston tap water. They then send those data to their other manufacturing facilities, which mimic the conditions to make beer that tastes the same as the beer in Boston.
This article was published on July 10, 2024.
Always free. No ads. Richly storied.
Always free. No ads. Richly storied.
Always free. No ads. Richly storied.