Stories We'd Like to Publish

Asimov Press has now existed for two months. (Woo!) We’re publishing an essay at a pace of roughly one per week, and our first printed compilation will be available in the next few weeks.
But there are only two of us (Niko & Xander), and our pile of essay ideas has already swelled to 450+ rows in a spreadsheet. At our current publishing cadence, it will take us at least nine years to tell all of these stories—and new ideas are being added every day. We’d like to launch more biotechnology stories into the universe, but we need your help to make that happen.
That’s why we are publicly sharing ideas for essays that we’d like to publish. If you’d like to pursue any of these, please email us: editors@asimov.com. We’re willing to work with both new and experienced writers; just let us know in your email why you might be the best person to write on the topic you’ve chosen. Examples of past work are also welcome. We’ll help you during every step of the writing process, from outline to drafts to editing. And then, to top it off, we’ll pay you, print your article in an upcoming essay collection, and invite you to attend one of our semi-annual launch parties.
Open-sourcing essay ideas is not an original idea. Our decision to do this was inspired by the Institute for Progress, an organization that publicly lists pieces that they’d “like to commission” or would “like to see out in the world.” It’s a great way to encourage readers to become writers. Arcadia Science does something similar with their Ice Box series, a collection of publicly accessible projects “that have been paused, or ‘put on ice.'"
At Asimov Press, our mission is to spread ideas that elucidate the promise of biotechnology, take its concomitant risks seriously, and direct talent toward solving pressing problems. As part of this mission, we’re seeking pieces that are mechanistic, quantitative, and nuanced. Stories should give enough detail to help readers understand which problems need to be solved and how we might arrive at solutions. We want this to be participatory, so please, let us know if there are any great ideas out there that we’ve missed.
{{signup}}
Story Ideas
The Making of Loyal
The longevity drug, LOY-001, is the first to receive “formal acceptance” from the F.D.A. Center for Veterinary Medicine for a drug “developed and approved to extend lifespan.” The startup behind it spent four years establishing a new regulatory path to bring lifespan extension drugs to market. But why has it historically been difficult to do this, and how can animal trials open up approvals for similar drugs for humans in the near future? What is the current state of the longevity drug market, and what do we know about how these drugs actually work?
Related:
- FDA Center for Veterinary Medicine agrees Loyal’s data supports reasonable expectation of effectiveness for large dog lifespan extension. Link
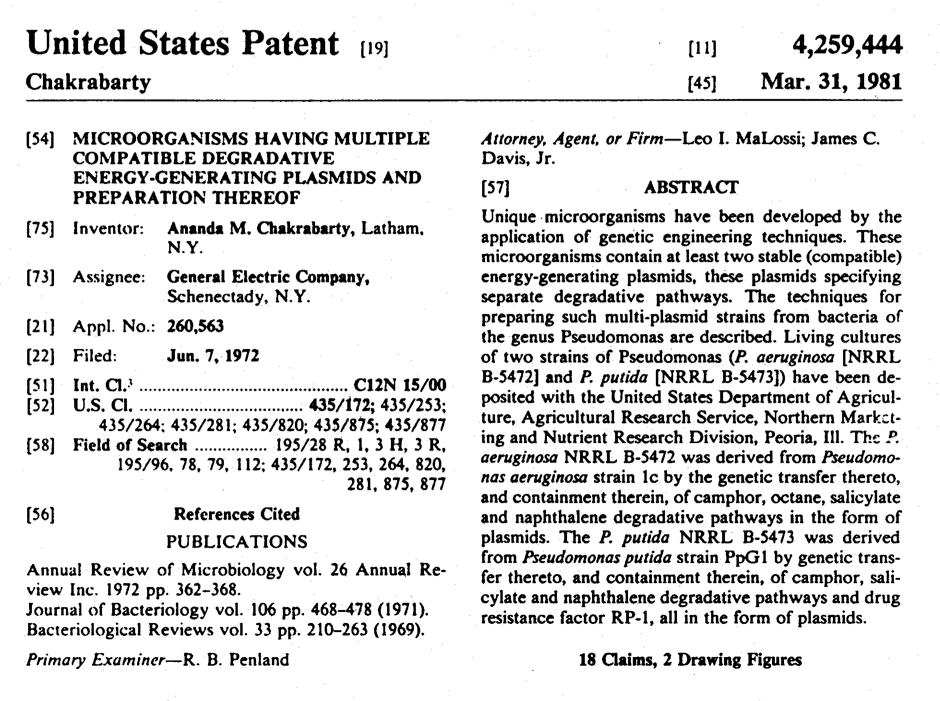
The First Patented GMOs
Ananda Chakrabarty, a genetic engineer at General Electric, was the first person to file a patent application for a genetically-altered organism in 1972 for a microbe engineered to clean up oil spills more rapidly than wildtype strains alone. We’d like to commission a brief history of this event and the court cases that followed. The article should clearly explain how the court decisions paved the way for the formal establishment of the biotechnology industry.
Related:
- Court Rules GE Can Patent Life Created in a Lab. Link
- Diamond v. Chakrabarty, 447 U.S. 303 (1980). Link
Where’s the Synthetic Blood?
Blood transfusions are widely needed in medical and surgical interventions. But we still rely on people to donate their own blood for others, which seems like a strangely out-of-date practice. Blood is difficult to store for long periods of time and hence needs constant donation, and some rarer blood types are often in short supply.
What have been the major scientific attempts to develop synthetic blood, and why they haven't succeeded yet? Thanks to Saloni Dattani for the recommendation.
Related:
- ABO gene editing for the conversion of blood type A to universal type O in Rhnull donor‐derived human‐induced pluripotent stem cells. Link
- Toward universal donor blood: Enzymatic conversion of A and B to O type. Link
- Artificial Blood Product One Step Closer to Reality With $46 Million in Federal Funding. Link
Biotechnology for Animal Welfare: Quantified
Animal suffering, particularly that which occurs as a result of factory-farming, is an issue that we’re certain future generations will look back on and say: Why did people allow this to continue? Unfortunately, it’s difficult to measure how many animals actually suffer and how much. But quantifying a problem is often the first step toward initiating impactful solutions.
More than 100 billion animals are killed for meat and other animal-derived products every year and there is a staggering number of wild animals that face disease, starvation, and predation in natural and urban environments. There have been attempts to quantify the number of animals in the wild, although this task is riddled with difficulties. 97 percent of living organisms are invertebrates and the estimated number of insects alone is 10 quintillion, with the majority of species still undiscovered.
Biotechnology offers many interventions against animal suffering.1 It has been used to spare cattle from the painful dehorning and to immuno-castrate pigs. It’s also used to sex chickens in-ovo which, while still a nascent technology, could save ~7 billion male chicks a year from maceration. We want to see more writing about what is possible in this space and what the bottlenecks are. While there is a role for fiction that imagines worlds in which animal agriculture is more humane, uncovering extant or near-term biotechnological solutions that are feasible, tractable, and economical is the way to actually bring about change.
Related:
- Animal Welfare Library. Link
- Animal Welfare - Rethink Priorities. Link
- Improving Genomic Selection for Heat Tolerance in Dairy Cattle: Current Opportunities and Future Directions. Link
- Biotechnology Could Change the Cattle Industry. Will it Succeed? Link
- A Scenario Analysis for Implementing Immunocastration as a Single Solution for Piglet Castration. Link
Real-Time Air Sensors
The COVID-19 pandemic highlighted a glaring hole in our diagnostic capabilities; namely, we’re too slow at—and need too much skilled labor—to diagnose sick people. Fortunately, new technologies could help to rapidly detect future outbreaks of influenza, coronaviruses, and other viral pathogens. In the last year, several papers have reported real-time surveillance technologies that detect pathogens in aerosols.
These devices, once installed in a home or hospital room, suck in air, collect pathogens into micro-wells, and then use sensitive biosensors and electronic systems to quantify circulating pathogens. We’d like to publish an in-depth look at how these new real-time surveillance tools work, where they fail, and what bottlenecks must be overcome to make them more widely accessible.
Related:
- Real-time environmental surveillance of SARS-CoV-2 aerosols. Link
- Air monitor detects airborne virus particles in real time. Link
- Towards Real-Time Airborne Pathogen Sensing. Link
The Challenge of Artificial Wombs
Artificial wombs simulate the uterus’ internal environment. They are already used, in some cases, to help premature babies born between 22 and 28 weeks to develop outside the womb. They have even broader ethical and social implications when it comes to the future of fertility. We’d like to publish a piece that elegantly explains how artificial wombs work and why it has been difficult to make technologies that can accommodate the full developmental span of an embryo, including before 22 weeks.
Related:
- Human trials of artificial wombs could start soon. Here’s what you need to know. Link
- Artificial womb technology and the frontiers of human reproduction: conceptual differences and potential implications. Link
Seeding the Universe
In 1948, John von Neumann described the Universal Assembler, a type of self-replicating machine. A Universal Assembler is a machine that can reproduce itself using resources in its local environment; a feat that living cells accomplish all the time. Could living cells be used to construct von Neumann probes—or self-replicating spacecrafts—that, when launched out into the Universe, could land on distant planets, replicate, and colonize their surfaces? We want to publish a piece that explains the current state of biological nanoprobes, how they work, and any technical or ethical challenges limiting their implementation.
Related:
- Picogram-Scale Interstellar Probes via Bioinspired Engineering. Link
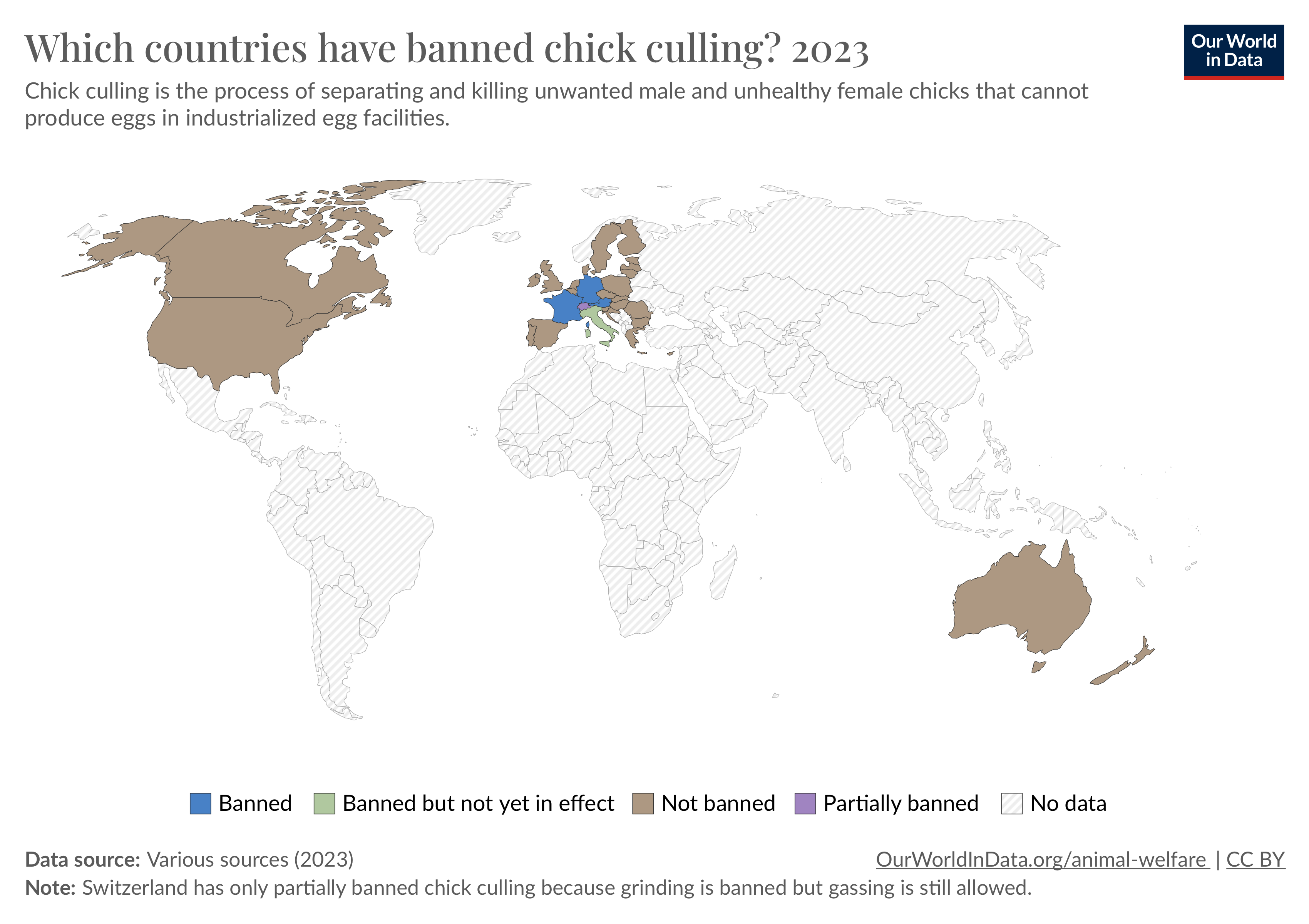
Biotechnology for Chick Culling
About 7 billion male chicks are culled each year in the egg industry. Males do not lay eggs, so the baby chicks are tossed into large blenders while still alive. Watch the videos for yourself if you dare. There is perhaps no greater opportunity for biotechnology to reduce superfluous animal pain and suffering than in creating solutions that eliminate chick culling. A deep dive article on this issue would explore in-ovo sexing, engineered chickens that lay eggs from which only females can hatch, and related technologies. The article should deeply explore economic and societal questions, too, including the question of why the egg industry (in the United States) hasn’t yet implemented technologies that already exist and are implemented elsewhere, and what it would cost them.
Related:
Speeding Up Cell Division
All applications in biotechnology flow downstream from engineered cells. So why is the field limited to working with organisms that only divide every 20 or 30 minutes (in the case of microbes) or every few hours (in the case of mammalian cells)? Some microbes, such as Vibrio natriegens, divide every ten minutes. Can we engineer organisms that divide even faster? What would be required to do that, what are the concomitant dangers, and to which applications could these ‘turbocharged’ cells be applied?
Related:
- Fundamental limits on the rate of bacterial growth and their influence on proteomic composition. Link
- Metabolic engineering of Vibrio natriegens. Link
DNA Assembly Bottlenecks
DNA synthesis has received a great deal of attention, and for good reason. In the 1960s, scientists invented solid-state synthesis methods to build short oligonucleotide sequences in a laborious, painstaking process. But in 2021, a company reported the first synthesis of a single strand of DNA that stretched more than 10,000 bases in length. In the next five years, DNA printers—machines that sit on a desk and physically print DNA molecules—will be able to synthesize DNA strands that stretch between five and seven thousand bases, according to estimates from the Nuclear Threat Initiative.
There has been far less attention devoted to DNA assembly. What is the most advanced way to stitch together lots of DNA sequences into, say, an entire genome? Why is this feat difficult, how might we make it possible to design, assemble and debug a full E. coli genome in one or two days, and how can we minimize the concomitant biosecurity risks?
Related:
- DNA synthesis technologies to close the gene writing gap. Link
- Benchtop DNA Synthesis Devices: Capabilities, Biosecurity Implications, and Governance. Link
Animal Birth Control
While surgical sterilization is the mainstay of pet population control, and poisons are the mainstay of pest population control, we are curious if we can do better. We’d like to publish a piece that examines reproductive control agents that have a biotechnology bent to them, including efforts in domestic cats and rodent populations. Just how feasible, expensive, and humane are they? And can they be applied more broadly?
Related:
- Durable contraception in the female domestic cat using viral-vectored delivery of a feline anti-Müllerian hormone transgene. Link
- The rodent birth control landscape. Link
DNA Screening Algorithms
Computer algorithms that sift through DNA sequences and detect possibly pathogenic sequences are an effective way to restrict access to would-be bioterrorists. But no federal mandates describe how these algorithms should work, exactly, or what sequences they must screen for. Many DNA synthesis providers basically “do their own thing,” although a recent Executive Order on Artificial Intelligence will mandate that researchers who receive federal funds only purchase DNA that has been screened. The same Order will also introduce “screening mechanisms, including standards and incentives.”
We don’t want to publish information hazards, but we do want to publish a piece that examines how DNA screening algorithms actually work and how they could be made better.
Related:
- Highlights of the 2023 Executive Order on Artificial Intelligence for Congress. Link
- Secure DNA. Link
- Preventing the Misuse of DNA Synthesis Technology. Link
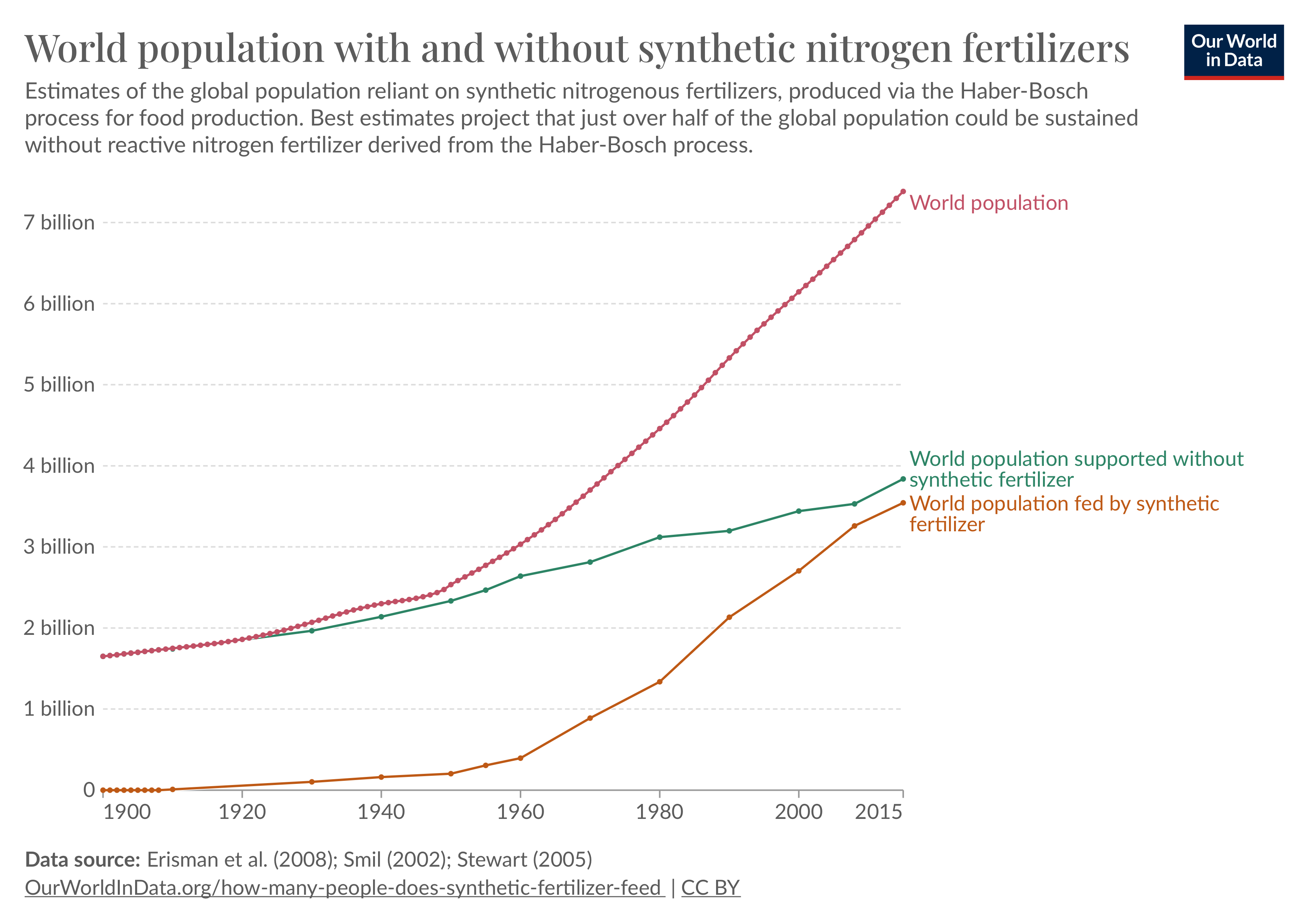
Synthetic Nitrogen Fixation
About 120 million metric tons of synthetic nitrogen fertilizers are made each year. Production of one ton of ammonia for such fertilizer, using natural gas, emits 1.6 tons of carbon dioxide. Farmers in the U.S. use about 72 kilograms of nitrogen fertilizer per hectare of land, according to Our World in Data, and there are 166 million hectares of cropland in the U.S. This means that the U.S. alone uses more than 13 million tons of fertilizer each year.
If plants could fix their own nitrogen—rather than rely on farmers’ use of synthetic fertilizers—we could slash global carbon emissions by billions of tons over a few years. The New Yorker recently ran an article on synthetic nitrogen fixation, but it did not cover alternative approaches to achieve the same ends, such as the use of engineered microbes that are planted with the seeds and provide nitrogen to the developing plant via symbiotic interactions (see Pivot Bio). We’d like to publish a mechanistic piece that examines synthetic nitrogen fixation, why it is difficult, and other strategies to curb our reliance on synthetic fertilizers.
Related:
Moore’s Law for Synthetic Gene Circuits
The first synthetic gene circuits—the toggle switch and repressilator—were made from just two or three genes, respectively. More recently, researchers have made synthetic gene circuits from 11 genes or, in another instance, from 62 genes spread out over 7 microbial strains. We’d like to publish a piece that traces the steadily growing size of synthetic gene circuits over time and explains why it may be difficult to scale them further. Why is it difficult to make large circuits that work as expected? Is it because we lack appropriate mathematical models, or because transgenes simply impose too much “burden” on the cells in which they are placed, or something else?
Related:
- Construction of a genetic toggle switch in Escherichia coli. Link
- A synthetic oscillatory network of transcriptional regulators. Link
- Genetic circuit design automation. Link
- Programming Escherichia coli to function as a digital display. Link
Making Pan-Vaccines
Current vaccines typically offer narrow protection against pathogens, such as influenza. As pathogens evolve and mutate, though, they can find ways to evade that protection, thus requiring that people get vaccinated repeatedly. In the last few years, dozens of research groups have begun developing pan-genus or pan-family virus vaccines. We’d like to publish a deep dive that explains how pan-vaccines for influenza actually work, how they are faring in clinical trials, and any challenges associated with their widespread rollout. A vaccine called M2e has been touted as a possible candidate for universal influenza protection for more than two decades now, but it requires “several administrations,” according to an article in PNAS, and does not effectively sustain “antibody titers over time.” What are the other options?
Related:
- A single-shot vaccine approach for the universal influenza A vaccine candidate M2e. Link
- Quadrivalent Influenza Vaccine. Link
- Mosaic RBD nanoparticles protect against challenge by diverse sarbecoviruses in animal models. Link
- Broad-spectrum pan-genus and pan-family virus vaccines. Link
The Difficulties with Tree Engineering
One-third of emitted carbon annually is captured and sequestered in forests. Trees are thus a promising avenue for carbon capture. Yet, Jack Wang, a tree engineer at North Carolina State University, estimates that there are fewer than 1,000 tree geneticists working today and only a few dozen people who actually know how to genetically engineer trees. We’d like to publish a piece that clearly explains why it is so challenging to engineer trees compared to crop plants and that highlights companies working in this space, such as TreeCo and Living Carbon.
Related:
- For the First Time, Genetically Modified Trees Have Been Planted in a U.S. Forest. Link
- Tree-Co. Link
- Living Carbon. Link
- Genetically edited wood could make paper more sustainable. Link
Costs of Cultivated Meat
The first burger made from cultivated meat debuted in 2013 and cost $330,000. Now, more than a decade later, this cost has fallen to about $9.80 a burger. A recent economic assessment suggests that the price of cultivated meat could come down to roughly $5.00 per kilo by 2030. But despite falling costs, pessimism about the future of cultivated meats abounds. Putting aside people’s feelings about broken promises and interindustry flubs, we want to publish a piece that explains how cultivated meat came down so far in price, and what exactly is limiting it from falling even further. This piece would be part history, part quantitative analysis.
Related:
- Is Cultivated Meat for Real? Link
- The Revolution That Died on Its Way to Dinner. Link
- Leveraging Biopharma Production Processes for Cultivated Meat. Link
- These Cities Aren’t Banning Meat. They Just Want You to Eat More Plants. Link
Key Biotechnology Numbers
Lists of numbers are a surprisingly effective way to help people think through ideas and identify obstacles. A friend of Asimov Press, Milan Cvitkovic, previously published a list of “Neurotechnology Numbers Worth Knowing.” He divided his list into sections about the brain, individual cells, hardware, and even operational concerns (such as: “A cage of 5 mice costs ~$1k upfront and ~$5k/yr recurring.”)
We’d like to publish similar lists of numbers, albeit for other facets of biotechnology. What numbers would be on a list of “Drug Numbers Worth Knowing” or “Synthetic Biology Numbers Worth Knowing”?
Related:
- Neurotechnology Numbers Worth Knowing. Link
The Protein Problem
Historically, most protein structures have been solved using x-ray crystallography, a technique in which proteins are removed from cells, packed into crystals, and then bombarded with x-ray beams. A sensor, placed behind the crystal, catches the diffracted x-rays, and the diffraction patterns are used to calculate the three-dimensional structure of the protein. This method erroneously portrays proteins as static, unmoving objects.
AlphaFold2, the computational model that predicts protein structures, was trained mostly on protein structures solved with x-ray crystallography. Proteins in cells behave more like liquids than solids, though; they wiggle to-and-fro in a chaotic dance and can adopt hundreds of distinct shapes. If one reverses AlphaFold’s predictions and instead makes the model generative, it tends to design proteins that are hyper-stable and rigid, much like the frozen proteins on which it was trained.
The question is: What technologies could enable one to “solve” protein structures—in all their wiggly variants—inside of living cells? We’re familiar with laboratories in Denmark that are working to develop in situ cryo-electron microscopy, a technique that would provide high-resolution structures of proteins without first pulling them from cells. We’d like to publish a piece that explains how these technologies work, and how they could make computationally-designed proteins more resemble the real things.
Related:
- Highly accurate protein structure prediction with AlphaFold. Link
- AlphaFold2 and its applications in the fields of biology and medicine. Link
- De novo protein design by inversion of the AlphaFold structure prediction network. Link
- ANewInSituCryo-ElectronMicroscopyApproachtoDirectly Visualize Mutations in Mitochondrial Disease. Link
- In situ cryo-electron tomography: a new method to elucidate cytoplasmic zoning at the molecular level. Link
Why Do Xenotransplants (Actually) Fail?
In January 2022, a 57-year-old man named David Bennett received an organ transplant: a heart from a genetically-edited pig. The pig heart was placed into Mr. Bennett’s chest and functioned normally for a time, but then everything went wrong. Bennett passed away that March.
At any given time, there are more than 100,000 people in the United States waiting on an organ transplant list. Organs harvested from engineered pigs could help to reduce this number, but xenotransplants have never worked in living people for an extended period of time. Pig-to-nonhuman primate transplants, however, have lasted >900 days for heart transplants and >400 days for kidney transplants. Although Bennett’s heart transplant likely failed because the organ was infected with a pig virus, according to an article in MIT Technology Review, we’d still like to know why xenotransplants have been so challenging to get right. An article should clearly explain how pigs are genetically altered to become organ donors, and what happens to a person’s immune system when pig organs are placed inside the body.
Related:
- A Brief History of Clinical Xenotransplantation. Link
- How Close Is Xenotransplantation, Really? Link
- Xenotransplantation: Past, Present, and Future. Link
- The Xenotransplant Patient Who Died Received a Heart Infected with a Pig Virus. Link
Predictive Growth Models
Theodor Escherich, an Austrian physician, was the first to isolate E. coli (from his own feces) in 1885. This simple microbe is now used as the organism-of-choice for nearly 15,000 biomedical research studies each year. Scientists tend to use E. coli because it is easy to work with and genetically tractable. But there are other microbes that grow in hydrothermal vents or survive in the vacuum of space—beautiful biology is just waiting to be discovered, if only we could grow and study such microbes in the laboratory.
We’d like to publish a piece that imagines how a predictive model, which infers the optimal growth nutrients for any microbe solely by looking at its genome sequence, could actually come to be. Such a sequence-to-growth model would vastly broaden the organisms that can be studied and might make it possible to concoct an “optimal broth” to grow a greater number of organisms. Small models can already tackle a modest version of this problem, but it’ll be a tall order to collect enough data to build a broadly general version. What datasets will be required? And how might biotechnology change as a result?
Related:
- What Biology Can Learn from Physics. Link
- Machine learning-assisted discovery of growth decision elements by relating bacterial population dynamics to environmental diversity. Link
{{divider}}
Footnotes
- Biotechnology in animal agriculture will not, and is not, only going to be used to promote positive welfare. Genetically engineering animals can help to improve animal agriculture productivity, increasing the rate of reproduction of domestic animals, and, with it, the quality and yield of animal production. While downstream effects such as promoting growth and improving nutrient intake efficiency may increase an animal’s welfare, the object here is to make animal husbandry cheaper and more efficient, thereby enabling its proliferation.
Always free. No ads. Richly storied.
Always free. No ads. Richly storied.
Always free. No ads. Richly storied.